RJPS Vol No: 15 Issue No: 3 eISSN: pISSN:2249-2208
Dear Authors,
We invite you to watch this comprehensive video guide on the process of submitting your article online. This video will provide you with step-by-step instructions to ensure a smooth and successful submission.
Thank you for your attention and cooperation.
1Department of Pharmaceutical Chemistry, Rani Chennamma College of Pharmacy, Belagavi, Karnataka, India
2Dr. Shrinivas Joshi, Novel Drug Design and Discovery Laboratory, Department of Pharmaceutical Chemistry, Soniya Education Trust’s College of Pharmacy, Sangolli Rayanna Nagar, Dharwad, Karnataka, India.
3Novel Drug Design and Discovery Laboratory, Department of Pharmaceutical Chemistry, Soniya Education Trust’s College of Pharmacy, Sangolli Rayanna Nagar, Dharwad, Karnataka, India
*Corresponding Author:
Dr. Shrinivas Joshi, Novel Drug Design and Discovery Laboratory, Department of Pharmaceutical Chemistry, Soniya Education Trust’s College of Pharmacy, Sangolli Rayanna Nagar, Dharwad, Karnataka, India., Email: shrinivasdj23@gmail.com
Abstract
Background: In Mycobacterium tuberculosis, the enzyme enoyl-acyl carrier protein (ACP) reductase (InhA) catalyzes the elongation of acyl fatty acids, which are essential building blocks of the mycobacterial cell wall and precursors of mycolic acids. Millions of people are at risk of contracting new TB infections, becoming ill, and dying, and a comprehensive understanding of the disease’s pathophysiology remains lacking.
Objectives: The objective was to develop potent therapeutic novel bioactive compounds that function as enoyl-acyl carrier protein reductase (InhA, PDB ID: 6R9W) inhibitors using an in silico drug design approach.
Methods: The virtual screening tool Auto Dock 1.5.6 was utilized to conduct molecular docking studies on enoyl-ACP reductase. Biovia Discovery Studio examined the docked complex of the ligand and protein.
Results: The ten bioactive compounds with the ability to bind to 6R9W exhibited docking scores ranging from -8.1 to -11.1 Kcal/mol. Compound 1, which showed the most favourable docking energy (-11.1 Kcal/mol), is an excellent candidate as a cell wall protein inhibitor (6R9W) and will be further investigated in vitro.
Conclusion: Compound 1, with the IUPAC name N'-((2-hydroxynaphthalen-1-yl) (phenyl)methyl)-4-(1Hpyrrol-1-yl) benzohydrazide, is an excellent candidate for the cell wall protein inhibitor (6R9W). It exhibited a strong docking affinity, with a binding energy of -11.1 kcal/mol. Using an in-silico drug design approach, the study highlights the effectiveness of pyrrole scaffolds as enoyl-acyl carrier protein (ACP) reductase (InhA) antagonists in novel antitubercular therapies.
Keywords
Downloads
-
1FullTextPDF
Article
Introduction
Tuberculosis (TB) is a highly dangerous infectious disease caused by the bacterial pathogen Mycobacterium tuberculosis (Mtb).1 It primarily affects the lungs, but can also involve other parts of the body. While uncommon in high-income countries, tuberculosis (TB) remains a significant public health issue, particularly in low- and middle-income populations. In 2012 alone, TB was the second leading cause of death from infectious diseases, claiming nearly 1.3 million lives-over 95% of which occurred in low- and middle-income nations.2 According to the most recent World Health Organisation (WHO) report, 8.6 million new cases of TB were reported, of which three million were co-infected with both HIV and Mycobacterium tuberculosis. 3 Individuals infected with tuberculosis relied heavily on a cocktail of 6-8 medications administered over extended periods, sometimes lasting up to 24 months.4 The majority of these medicines have several limitations, including host toxicity and ineffectiveness against multidrug-resistant TB (MDR-TB).5 Therefore, there is a greater need than ever for new and efficient chemotherapeutic medicines to shorten the course of treatment and minimise adverse effects in order to fight this illness.6
Pyrrole and its derivatives are key nitrogen-containing heterocyclic compounds present in a wide range of natural products, such as bile pigments, heme, cytochromes, vitamin B12, and alkaloids. One of its many additional qualities is antimycobacterial action.7,8
Molecular docking is a strategy for determining how one molecule should interact with another when they are combined to produce a stable combination.9 Using specific characteristics, one can predict the degree of interaction or ligands between two molecules based on their preferred orientation. Scoring techniques are used to assess the strength of the non-covalent contact, sometimes referred to as binding affinity, between two molecules when they are docked.10 Molecular docking is an increasingly valuable tool in drug discovery.
The enoyl-acyl carrier protein (ACP) reductase (InhA) of Mycobacterium tuberculosis has emerged as a promising target for developing antitubercular drugs.11 InhA belongs to the family of NADH-dependent enoyl-ACP (CoA) reductase enzymes. Acyl fatty acids, mycolic acid precursors, and components of mycobacterial cell walls are all expanded by InhA. This study used a structure-based computer simulation of the relationships between ligands and receptors. The aim of this work was to identify potentially bioactive compounds that serve as enoyl acyl carrier protein (ACP) reductase (InhA) antagonists using an in silico drug discovery approach.
Materials and Methods
Virtual AutoDock Tool version 1.5.6 was utilized to perform the in silico molecular docking analysis. The protein was prepared using Biovia Discovery Studio software version 2021. Auto Dock Tool 1.5.6. was used for ligand preparation, grid creation, and receptor-ligand docking. The SwissADME server was used to predict drug-likeness for all compounds included in the investigation.
Selection and preparation of target protein
The enoyl-acyl carrier protein (ACP) reductase (InhA), PDB ID: 6R9W, was obtained from the protein data bank (PDB) with 1.75 resolution for docking. The protein was prepared for molecular docking using Biovia Discovery Studio software version 2021. Initially, all non-essential elements, encompassing heteroatoms like water molecules and co-crystallized ligands were eliminated. Following this, the protein's structure was enhanced by the addition of hydrogen atoms to polar groups, and Gasteiger charges were computed to account for electrostatic attributes.
Selection of ligand molecules
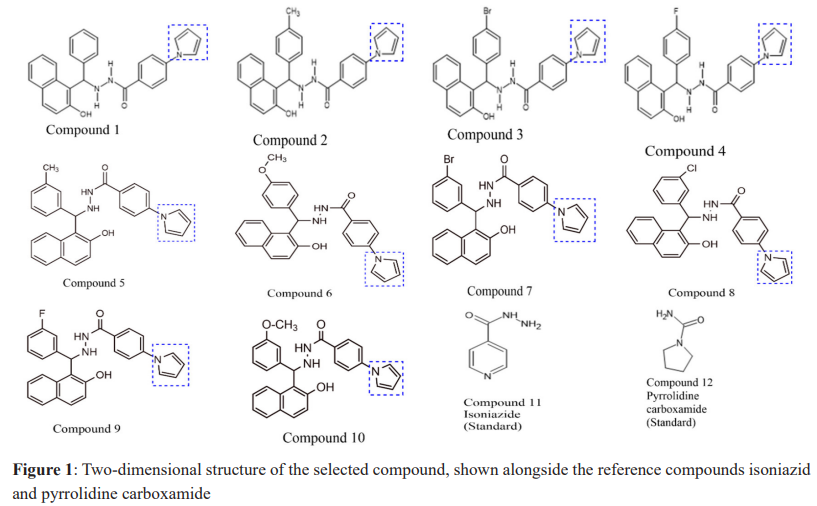
For the selection of ligand molecules, a target-based ligand selection approach was employed. The compounds that demonstrated the best interaction and docking score were chosen, and were synthesized. They were manually created using ChemSketch as depicted in Figure 1. The Open Babel software played a pivotal role in seamlessly converting various file formats essential for our research.12 Furthermore, for the critical task of energy minimization of the ligand,13 Argus Lab, a software package specializing in molecular modeling, graphical visualization, and drug design was employed. To filter all bioactive compounds based on the Lipinski Rule of Five, additional ADME predictions were performed for all structures using SwissADME.14
Molecular docking investigation
The molecular docking study was performed using AutoDock Tool version 1.5.6. The prepared protein structure was uploaded as a macromolecule, and bioactive chemicals (ligands) were selected. Prior to converting it to Auto Dock ligand format (pdbqt), the software first decreased the ligand energy. After using the grid box to cover the complete protein structure, blind docking was finally started to identify the bioactive chemicals that best fit the energy value. There were roughly ten runs of each simulation, producing ten docked conformations. Consequently, it was demonstrated that the strongest binding conformations were found in configurations with the lowest energy. Finally, Biovia Discovery Studio examined the optimal ligand and protein docked combination.15
Results
The 6R9W structure of enoyl-[acyl-carrier-protein] reductase (NADH, comprising ~272 amino acids, has been elucidated at a resolution of 1.75 Å. In silico molecular testing was performed on many bioactive substances. The number of rotatable bonds, molecular weight, hydrogen bond donor and acceptor, and other physiochemical properties were anticipated and examined for drug infractions. All of these compounds adhered to the "Lipinski Rule of Five" as shown in Table 1.
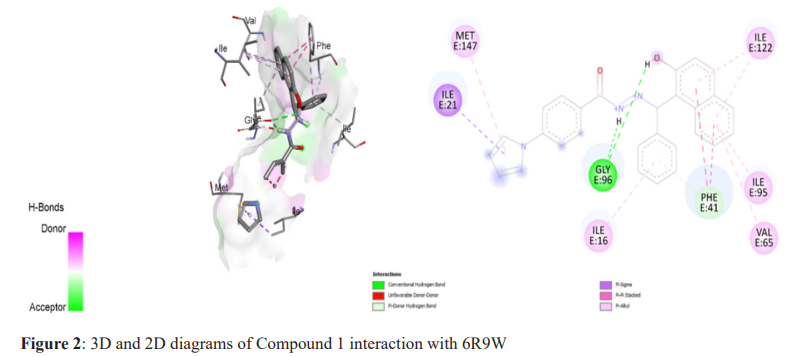
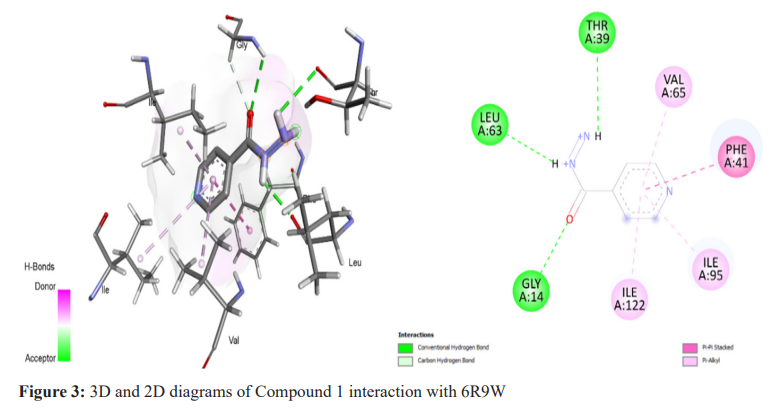
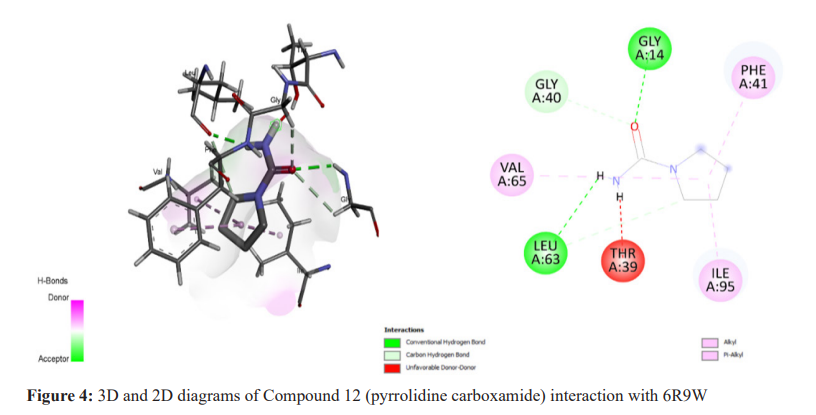
The molecular docking technique was used to investigate, predict, and comprehend the protein or enzyme interactions with the specified ligand library, and to visualise the likely binding. Pyrrole derivatives were docked with receptors (6R9W) using Biovia Discovery Studio, the virtual screening tool which used Autodock Tool 1.5.6. Binding energies and interacting residues were discovered and are summarized in Table 2. The selected compound with the highest docking energy (Compound 1) and the reference compounds are illustrated in 3D and 2D diagrams with protein 6R9W (Figure 2-4).
Discussion
Tuberculosis is highly contagious and spreads through aerosols, posing particular risk to infants and individuals with impaired immune systems. Mycobacterium tuberculosis can persist asymptomatically for years if macrophages fail to eliminate the pathogen.16 Reactivation may occur at any time, and pulmonary tuberculosis is observed in approximately 80% of cases. The need for effective treatment has prompted researchers to develop and synthesize new substances capable of targeting both fungal and mycobacterial infections. The emergence of antibiotic-resistant strains has made the search for medications with potent antitubercular activity increasingly crucial in tuberculosis research.17 The objective of this study was to develop compounds capable of binding to the 6R9W protein, with a potential to serve as future therapeutics against Mycobacterium tuberculosis. Approximately ten bioactive molecules were identified as binding to 6R9W, with binding energy ranging from -8.1 to -11.1 Kcal/moL.
Compound 1, with the IUPAC name N'-((2-hydroxynaphthalen-1-yl) (phenyl)methyl)-4-(1H-pyrrol-1-yl) benzohydrazide, was identified as the best ligand, exhibiting the lowest binding energy (-11.1 Kcal/mol) with 6R9W (Figure 2, Table 2). During the global Mycobacterium tuberculosis outbreak, isoniazid and pyrrolidine carboxamides were used as the therapeutic agents, along with numerous antiviral compounds (Figure 3, 4 and Table 2). An effort was made to predict these drugs’ binding mechanisms and binding energies with the 6R9W protein, showing binding energies of -9.9 Kcal/mol for isoniazid and -8.1 Kcal/mol for pyrrolidine carboxamide. Furthermore, all compounds were observed to restrict viral reproduction and translation, which may significantly influence viral load reduction in infected patients.18,19 In comparison to pyrrolidine carboxamide and isoniazid, all the compounds tested here had a higher negative binding potential energy. All bioactive substances utilized in this investigation complied with Lipinski’s Rule of Five, indicating their potential for therapeutic use (Table 1). Our findings are consistent with previous reports on isoniazid derivatives and their antitubercular action,20 as well as on pyrrolidine carboxamide as a new class of enoyl acyl carrier protein inhibitors.21
Chemical libraries can be compared to possible drug candidates using Biovia Discovery Studio, a virtual screening tool for computational therapeutic discovery. Biovia Discovery Studio walks users through the whole process, from data pre-processing to job submission and assessment, and enables medicinal chemists to conduct virtual screening from any place. Biovia Discovery Studio's docking method, which emphasizes simplicity, is beneficial to computer-aided pharmaceutical research, despite the fact that there is no magic solution in drug discovery.
Conclusion
Docking of molecules is part of a revolutionary novel approach for studying the binding of small compounds to receptor proteins. The antituberculosis efficacy of pyrrole scaffolds as enoyl-acyl carrier protein (ACP) reductase (InhA) inhibitors has been explored in novel therapies. Compound 1, with the IUPAC name N'- ((2-hydroxynaphthalen-1-yl) (phenyl)methyl)-4-(1Hpyrrol-1-yl) benzohydrazide, was found to exhibit the most favourable docking energy (-11.1Kcal/mol) and is thus considered a potential inhibitor of cell wall proteins (6R9W). Further in vitro investigation is recommended.
Conflicts of Interest
The authors declare no conflict of interest.
Supporting File
References
- Rothschild BM, Martin LD, Lev G, et al. Mycobacterium tuberculosis complex DNA from an extinct bison dated 17,000 years before the present. Clin Infect Dis 2001;33(3):305-11.
- Tollefson D, Bloss E, Fanning A, et al. Burden of tuberculosis in indigenous peoples globally: a systematic review. Int J Tuberc Lung Dis 2013;17(9):1139-50.
- Vladimirova S, Bijev A. Synthesis of new derivatives of pyrrole tuberculostatics providing structural diversity. Pharmacia 2015;62(1):3-8.
- Stephanie F, Saragih M, Tambunan US. Recent progress and challenges for drug-resistant tuberculosis treatment. Pharmaceutics 2021;13(5):592.
- Surineni G, Yogeeswari P, SriramD, et al. Design and synthesis of novel carbazole tethered pyrrole derivatives as potent inhibitors of Mycobacterium tuberculosis. Bioorg Med Chem Lett 2015;25(3):485-91.
- Zumla AI, Gillespie SH, Hoelscher M, et al. New antituberculosis drugs, regimens, and adjunct therapies: needs, advances, and future prospects. Lancet Infect Dis 2014;14(4):327-40.
- Gholap SS. Pyrrole: An emerging scaffold for construction of valuable therapeutic agents. Eur J Med Chem 2016;110:13-31.
- Fan H, Peng J, HamannMT, et al. Lamellarins and related pyrrole-derived alkaloids from marine organisms. Chem Rev 2008;108(1):264-87.
- Jiao X, Jin X, Ma Y, et al. A comprehensive application: Molecular docking and network pharmacology for the prediction of bioactive constituents and elucidation of mechanisms of action in component-based Chinese medicine. Comput Biol Chem 2021;90:107402.
- Thomas L, Birangal SR, Ray R, et al. Prediction of potential drug interactions between repurposed COVID-19 and antitubercular drugs: an integrational approach of drug information software and computational techniques data. Ther Adv Drug Saf 2021;12:20420986-211041277.
- Dogan SD, Gunduz MG, DoganH, et al. Design and synthesis of thiourea-based derivatives as Mycobacterium tuberculosis growth and enoyl acyl carrier protein reductase (InhA) inhibitors. Eur J Med Chem 2020;199:112402.
- Boyle NM, Banck M, James CA, et al. Open Babel: An open chemical toolbox. J Cheminform 2011;3:1-4.
- Thompson M. Argus Lab: Molecular modeling, graphics, and drug design program [Internet]. Seattle (WA): Mark Thompson. 2025 Sep 20. Available from:http://www.arguslab.com/arguslab.com/ ArgusLab.html.
- Parmar GR, Shah AP, Sailor GU, et al. In Silico discovery of novel phytoconstituents of amyrispinnata as a mitotic spindle kinase inhibitor. Current Drug Research Reviews Formerly: Curr Drug Abuse Rev 2020;12(2):175-82.
- Noh MA, Fazalul Rahiman SS, A Wahab H, et al. Discovery of new targeting agents against GAPDH receptor for antituberculosis drug delivery. J Basic Clin Physiol Pharmacol 2021;32(4):715-22.
- Seki M, Choi H, Kim K, et al. Tuberculosis: A persistent unpleasant neighbour of humans. J Infect Public Heal 2021;14(4):508-13.
- Radan M, Ruzic D, AntonijevicM, et al. In silico identification of novel 5-HT2A antagonists supported with ligand-and target-based drug design methodologies. J Biomol Struct Dyn 2021;39(5):1819-37.
- Arshad A, Dayal S, Gadhe R, et al. Analysis of tuberculosis meningitis pathogenesis, diagnosis, and treatment. J Clin Med 2020;9(9):2962.
- Stelitano G, Sammartino JC, Chiarelli LR. Multitargeting compounds: a promising strategy to overcome multi-drug-resistant tuberculosis. Molecules 2020;25(5):1239.
- Kalani K, Chaturvedi V, Trivedi P, et al. Dihydroartemisinin and its analogs: a new class of antitubercular agents. Curr Top Med Chem 2019;19(8):594-9.
- Prasad MS, Bhole RP, Khedekar PB, et al. Mycobacterium enoyl acyl carrier protein reductase (InhA): A key target for antitubercular drug discovery. Bioorg Chem 2021;115:105242.